Eukaryotic gene expression without intervention by a Morpholino
In
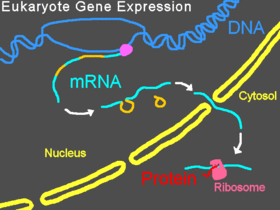
eukaryotic organisms, pre-mRNA is
transcribed in the nucleus,
introns are
spliced out, then the mature mRNA is exported from the
nucleus to the
cytoplasm. The small subunit of the
ribosome usually starts by binding to one end of the mRNA and is joined there by various other
eukaryotic initiation factors, forming the initiation complex. The initiation complex scans along the mRNA strand until it reaches a
start codon, and then the large subunit of the ribosome attaches to the small subunit and
translation of a
protein begins. This entire process is referred to as gene expression; it is the process by which the information in a
gene, encoded as a sequence of bases in
DNA,
is converted into the structure of a protein. A Morpholino can modify
splicing or block translation, depending on the Morpholino's base
sequence.
Blocking translation
Translation blocked by a Morpholino oligo
Bound to the
5'-untranslated region of messenger RNA (mRNA), Morpholinos can interfere with progression of the
ribosomal initiation complex from the 5' cap to the start codon. This prevents
translation of the coding region of the targeted
transcript (called "
knocking down"
gene expression).
This is useful experimentally when an investigator wishes to know the
function of a particular protein; Morpholinos provide a convenient means
of knocking down expression of the protein and learning how that
knockdown changes the cells or organism. Some Morpholinos knock down
expression so effectively that, after degradation of preexisting
proteins, the targeted proteins become undetectable by
Western blot.
[16]
In 2016 a synthetic peptide-conjugated PMO (PPMO) was found to inhibit the expression of
New Delhi Metallo-beta-lactamase, an enzyme that many drug-resistant bacteria use to destroy carbapenems.
[17][18]
Modifying pre-mRNA splicing
Splicing blocked by a Morpholino oligo
Morpholinos can interfere with
pre-mRNA processing steps either by preventing splice-directing small nuclear ribonucleoproteins (
snRNP) complexes from binding to their targets at the borders of introns on a strand of pre-mRNA, or by blocking the
nucleophilic adenine
base and preventing it from forming the splice lariat structure, or by
interfering with the binding of splice regulatory proteins such as
splice silencers
[19] and
splice enhancers.
[20] Preventing the binding of snRNP
U1 (at the donor site) or
U2/
U5 (at the polypyrimidine moiety and acceptor site) can cause modified
splicing, commonly excluding
exons
from the mature mRNA. Targeting some splice targets results in intron
inclusions, while activation of cryptic splice sites can lead to partial
inclusions or exclusions.
[21] Targets of
U11/
U12 snRNPs can also be blocked.
[22] Splice modification can be conveniently assayed by reverse-transcriptase polymerase chain reaction (
RT-PCR) and is seen as a band shift after
gel electrophoresis of RT-PCR products.
[3]


No comments:
Post a Comment